New study identifies one mechanism that helps cancer cells withstand genetic mutations
Friday, 13 June, 2025
Share
Congratulations to Associate Professor Colm Ryan and collaborators across UCD Conway Institute, UCD Systems Biology Ireland and University of Edinburgh on their recently published research in Molecular Systems Biology titled 'Proteomic compensation by paralogs preserves protein interaction networks after gene loss in cancer'.
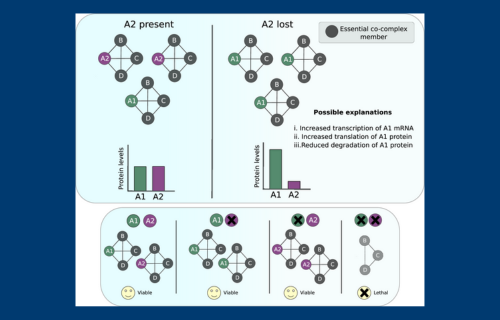
One remarkable aspect of cancer cells is their ability to cope with extensive DNA mutations. Even though many genes in cancer cells become damaged or nonfunctional due to these mutations, cancer cells typically grow faster than healthy cells. This resilience contrasts sharply with human-designed systems: if you removed just a few components from your phone, it would quickly stop working. Our study identifies one mechanism that helps cancer cells withstand genetic mutations: the use of "backup" or duplicate genes. These backup genes can perform similar roles to the damaged genes, essentially acting as spare copies. We find that often when one gene is lost or mutated in cancer, the protein produced by a backup gene is increased, compensating for the lost gene. Targeting these backup systems might therefore provide a powerful strategy to selectively kill cancer cells and develop more effective cancer treatments.
Abstract: Proteins operate within dense interconnected networks, with interactions necessary both for stabilising proteins and enabling them to execute their molecular functions. Remarkably, protein–protein interaction networks operating within tumour cells continue to function despite widespread genetic perturbations. Previous work has demonstrated that tumour cells tolerate perturbations of paralogs better than perturbations of singleton genes, but the underlying mechanisms remain poorly understood. Here, we systematically profile the proteomic response of tumours and cell lines to gene loss. We find many examples of proteomic compensation, where loss of one gene causes increased abundance of a paralog, and collateral loss, where gene loss causes reduced paralog abundance. Compensation is enriched among paralog pairs that are central in the protein–protein interaction network and whose interaction partners perform essential functions. Compensation is also significantly more likely to be observed between synthetic lethal pairs. Our results support a model whereby loss of one gene results in increased protein abundance of its paralog, stabilising the protein–protein interaction network. Consequently, tumour cells may become dependent on the paralog for survival, creating potentially targetable vulnerabilities.
See the full paper in Molecular Systems Biology (opens in a new window)here.