New Prognostic Marker in Childhood and Adolescent Leukaemia
Leukaemia is the most common paediatric cancer. Although most children and adolescents are completely cured of their disease, some don’t do so well. Also, the treatments that are currently used for leukaemias can occasionally have severe side-effects. We need to have a better understanding of the biology of the leukaemia cells in order to improve treatments and outcomes for these patients.
Some children and adolescents with acute myeloid leukaemia (AML) have a poor prognosis, but it is not always clear why. Sometimes this relates to the types of genetic mutations that are present in the leukaemia cells. If we can identify cases with poor-risk mutations early on, it means we can tailor their treatment accordingly. This can also give us clues about the biology of the leukaemia, and can help us to investigate more targeted treatments for the disease.
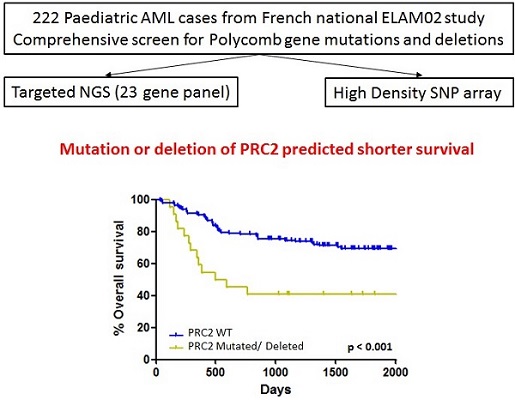
We have discovered a new predictor of prognosis in childhood AML. We did a targeted genetic screen, and found that mutations and deletions of genes that code for a family of proteins, known as the Polycomb protein complex PRC2, were strongly associated with a poor outcome.
Polycomb factors are interesting proteins. They work as part of protein complexes and help to control how lots of other genes are expressed. If Polycomb genes are mutated or deleted, this is likely to affect the levels of lots of other genes. Part of my research program in SBI will investigate the precise mechanisms by which Polycomb loss changes the biology of the leukaemia, and how this might influence treatment outcome. This is the first time that mutations in Polycomb genes have been found to predict outcome in any blood cancer. This is also a brand new specific prognostic marker in childhood AML.
The genetic screen comprised two approaches:
1) A targeted next generation sequencing (NGS) panel to test for mutations in Polycomb genes. This was carried out in my previous laboratory in Hôpital Necker-Enfants Malades in Paris.
2) A Single nucleotide polymorphism (SNP) array to test for gene deletions. SNPs are the most common type of genetic variation among people. This was carried out by our collaborators in Lille.
We then tested whether alterations in any specific genes predicted outcome in childhood AML, in collaboration with our colleagues at the Hôpital Trousseau in Paris. We found that grouping the genes by their function gave us more information than looking at any one gene alone. Specifically in this case, loss of any of the genes of the core Polycomb repressive complex 2 (PRC2) complex (EZH2, SUZ12 and EED) correlated with shorter survival.
Publication Reference:
Polycomb repressive complex 2 haploinsufficiency identifies a high-risk subgroup of pediatric acute myeloid leukemia. Bond J, Labis E, Marceau-Renaut A, Duployez N, Labopin M, Hypolite G, Michel G, Ducassou S, Boutroux H, Nelken B, Bertrand Y, Baruchel A, Petit A, Asnafi V, Leverger G, Preudhomme C, Macintyre E, Lapillonne H. Leukemia. 2018 Jun 27. doi: 10.1038/s41375-018-0187-9.
About the author:

Jonathan Bond is a Professor of Paediatric Molecular Haemato-Oncology and is leading a new long-term collaborative paediatric leukaemia research program between Our Lady’s Children’s Hospital Crumlin and Systems Biology Ireland at UCD, supported by the National Children’s Research Centre (NCRC), Crumlin. His research focusses on how gene regulation gets disrupted in leukaemia.

